In 1857, a young pediatrician named Theodor Escherich discovered what may very well be the most well-studied organism today. The rod-shaped bacterium named Escherichia coli, better known as E. coli, is a very common microbe residing in your gut. It’s also the workhorse of early molecular biology.
Luck likely played a role in its rise in popularity among scientists. Even under 19th-century lab conditions, where sterilization techniques were not perfect and little was known about what food bacteria need to survive, this microbe was easy to cultivate and grow quickly. It can replicate in under 20 minutes and can use a variety of carbon sources for energy.
As the first species to have its physiology thoroughly explored, E. coli has contributed fundamental knowledge to the fields of microbiology, molecular genetics and biochemistry, including how DNA replicates, how genes create proteins and how bacteria share genetic material among themselves – a huge cause of antibiotic resistance.
VectorMine/iStock via Getty Images Plus
However, the favored use of E. coli in the lab has also led to oversimplifications in the world of microbiology, distracting researchers from the thousands of other bacterial species that remain understudied.
As microbiologists studying the inner mechanisms of antibiotic tolerance, we and colleagues in our lab examine bacterial species that physiologically differ from E. coli in hopes of expanding the existing pool of knowledge within microbiology. For instance, drugs like penicillin fall into a class of antibiotics that target the outer defenses of the bacteria. We found that while E. coli succumbs to this attack, species like Vibrio or Klebsiella can tolerate it and survive.
A one-size-fits-all approach may have worked in the past, but embracing the true diversity of microbes could help scientists better fight the rise of antibiotic resistance.
Scientific good of E. coli
Researchers worked out the very foundations of life using E. coli. The significance of this bacterium for the field of biology is probably best captured by the biochemist Jacques Monod, who famously said, “What is true for E. coli is true for the elephant.”
Because researchers were able to watch regions of E. coli‘s DNA become mobile, allowing bacteria to transfer DNA among one another in a process called conjugation, scientists learned to manipulate this process to genetically alter organisms and study the effects of different genes.
E. coli helped reveal that bacterial chromosomes are circular and that manipulating a specific enzyme can allow scientists to easily clone parts of the bacterial genome.
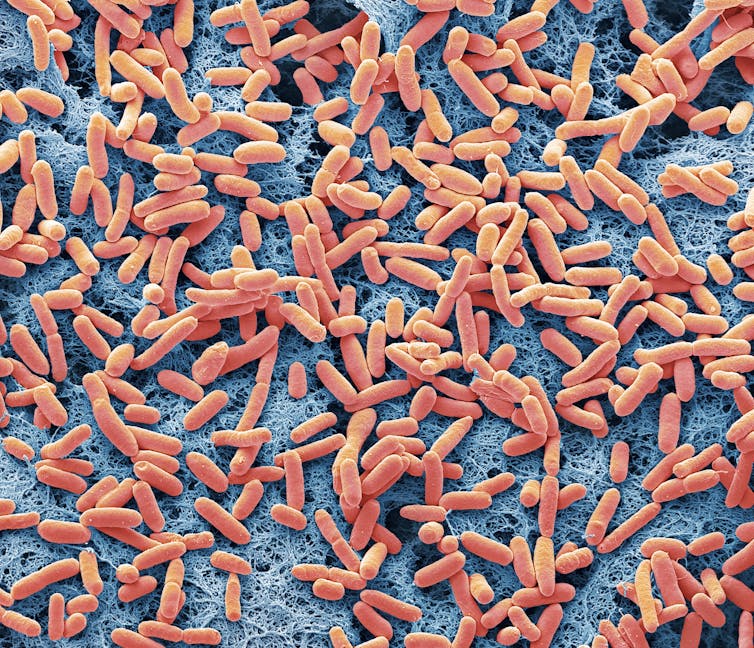
Steve Gschmeissner/Science Photo Library via Getty Images
E. coli also opened doors to using a type of bacterial viruses called phages as an alternative to antibiotics.
Widely available knowledge about and methods to study E. coli led to its prominence in academic and commercial research and drug production. In 2015, nearly 30% of proteins used as treatments for a wide range of diseases like hepatitis C and multiple sclerosis were derived from E. coli.
Model organism drawbacks
E. coli’s track record has solidified its place in the lab as a model organism. Model organisms are nonhuman species researchers use to study biology, with the expectation that the findings can be applied to other species like humans. Species are often chosen for their ease of maintenance, quick life cycles and overall cost-effectiveness.
However, model organisms have their drawbacks. Some researchers have argued that drawing parallels across species can sometimes fall short, leading to assumptions about more complex species that may not be true.
Additionally, study findings using nonmodel organisms are often less visible in the broader scientific community, since many researchers focus on organisms with known and defined traits. This bias results in a shadow space where progress is not immediately incorporated into broader scientific knowledge, which can slow down research that actually covers a range from bacteria to elephants.
ESKAPE pathogens don’t include E. coli
Model organisms are not perfect, and E. coli may not be an effective species to use to study many human bacterial infections. Focusing research on this microbe limits the exploration of how other bacteria infiltrate and infect human hosts. While some strains of E. coli can be deadly, they are not the only worrisome pathogens today.
ESKAPE pathogens, a group of bacteria that are highly resistant to antibiotics, pose a massive global health threat because they can quickly evolve traits that allow them to evade immune systems and available treatments. Species within ESKAPE, such as Klebsiella pneumoniae and E. cloacae, are able to resist multiple drugs and exhibit physical characteristics that E. coli does not, such as the ability to remove their cell wall and evade certain drugs.
Our lab is studying the unique traits that allow ESKAPE pathogens to survive antibiotics – traits we would not have known about if we used only E. coli as a model organism in our research.
With the many basics of fundamental bacterial cell and molecular biology covered thanks to E. coli, it may be time for researchers to turn toward the new pathogens wreaking havoc on society. Model organisms are wondrous tools, but they have limited power to allow findings to be extrapolated to other organisms. Better understanding the underpinnings of bacterial infections and antibiotics for a given disease requires studying the specific organism.




